
home > research > structural and dynamic studies of NA
Structural and dynamic studies of NA
(project within SFB 902: Structure and conformational flexibility of RNA studied by EPR spectroscopy)This project aims to investigate structural and conformational flexibility aspects of RNA and RNA-protein complexes by continuous-wave (cw) EPR and Pulsed Electron-Electron Double Resonance (PELDOR) spectroscopy performed at several magnetic field values (from 0.1 to 6.4 T). Spin-labeled RNA is used to explore structure, conformational flexibility and dynamics of tertiary folded RNA motives as well as the structural transitions induced by binding to proteins, metal ions or ligands. Our preliminary work on nucleic acid molecules is summarized in the following book chapters. [1]
Quantitative characterization of small RNA motives
We investigate structure and conformational flexibility of several small RNA motives with different three-dimensional architecture and compare
our multi-frequency, multi-field PELDOR analysis with data from NMR, X-ray, FRET and MD studies on the same systems to evaluate the potential of
the PELDOR method in this respect. The question to be answered is, whether it is possible to get a unique solution of the conformational flexibility
of such RNA motives. Furthermore we ask if the conformational flexibility determined by PELDOR at low temperatures can be related to the conformational
dynamics occur at physiological conditions.
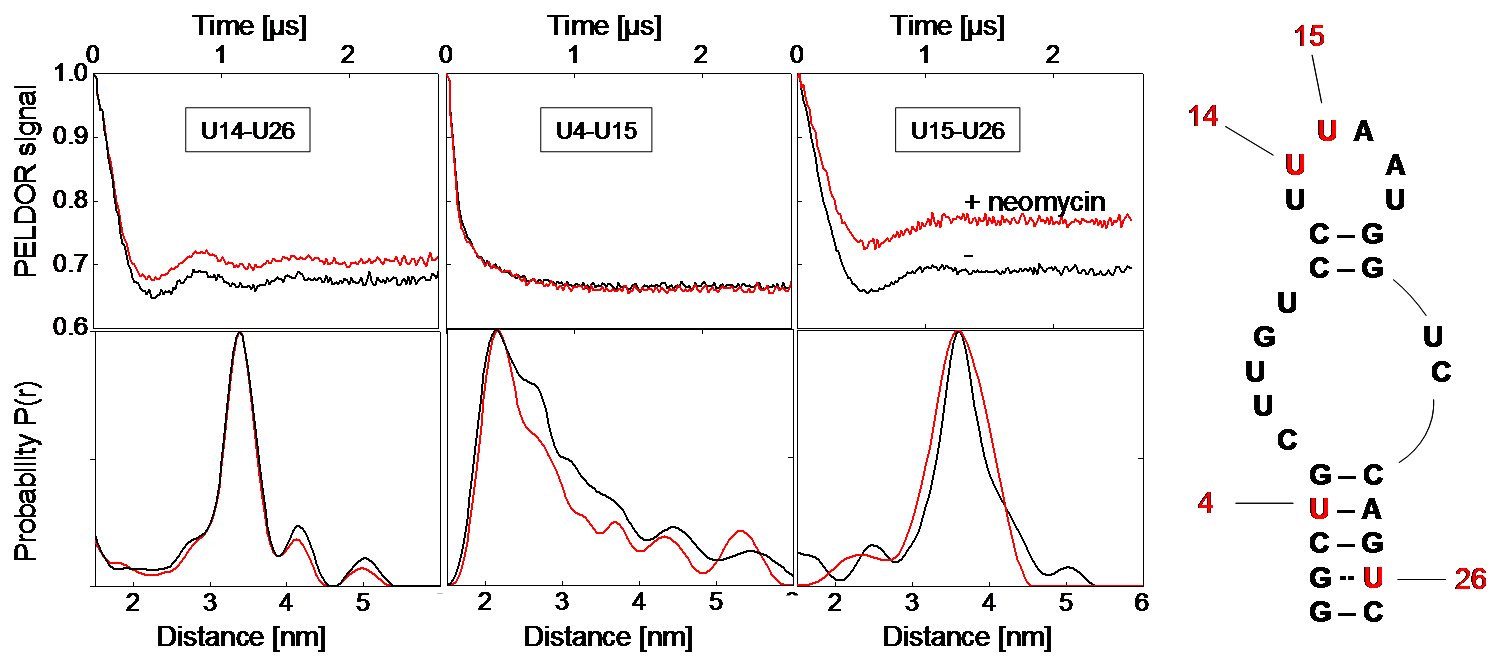
Left: Background-corrected PELDOR time traces and corresponding distance distributions of the three double-labeled neomycin-responsive riboswitch samples in presence (red) and absence of neomycin (black). Right: Secondary structure of the neomycin-responsive riboswitch with positions of spin-labeled nucleotides. [2]
Evaluation of spin-labeling strategies for long RNA molecules and in-cell applications
The development of spin-labeling techniques for RNA molecules is one of our main issues. The long-term goal is to explore the possibility of applying PELDOR under physiological conditions (room temperature, in-cell) to large RNAs or RNA/protein complexes. Aim of the project is to characterize the structure of short nucleic acid duplexes, dsRNA as well as dsDNA, inside cells; this will shed light on how the native environment affects the conformation of the studied systems with respect to the findings obtained in buffered solutions. In the current stage of the project the investigation is performed in Xenopus lævis oocytes. The structural information is obtained from pulse dipolar spectroscopy experiments, mainly PELDOR, performed on nucleic acid molecules which are doubly labelled with newly developed spin labels; these latter are designed to provide accurate conformational restraints and to withstand the reducing conditions characterizing the intracellular environment. Due to the characteristic of the investigated samples, the advancement of the project is in tight connection with the development of high-sensitivity techniques in EPR.
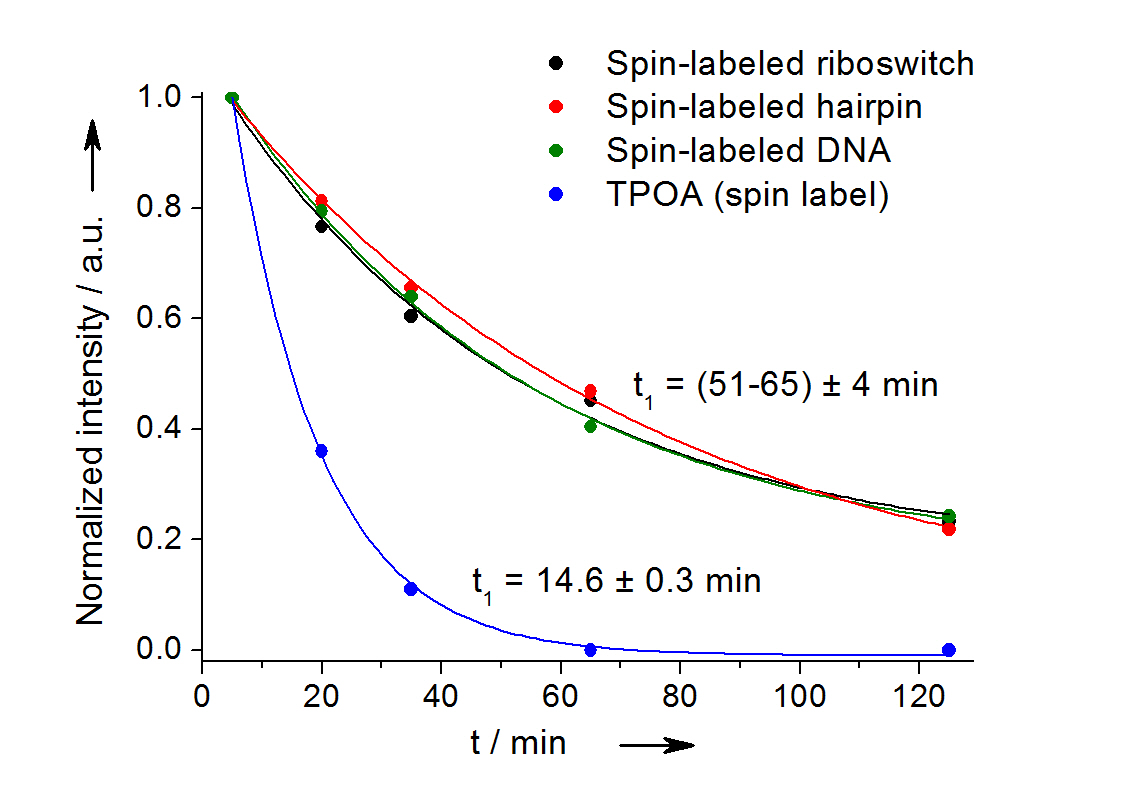
In-cell reduction kinetic curves for the spin label nitroxide attached to a 27-mer neomycin-sensing riboswitch (black), a 14-mer RNA hairpin (red), a 12-bp duplex DNA (green) and for the free spin label (blue). [3]
Hyperfine Spectroscopy
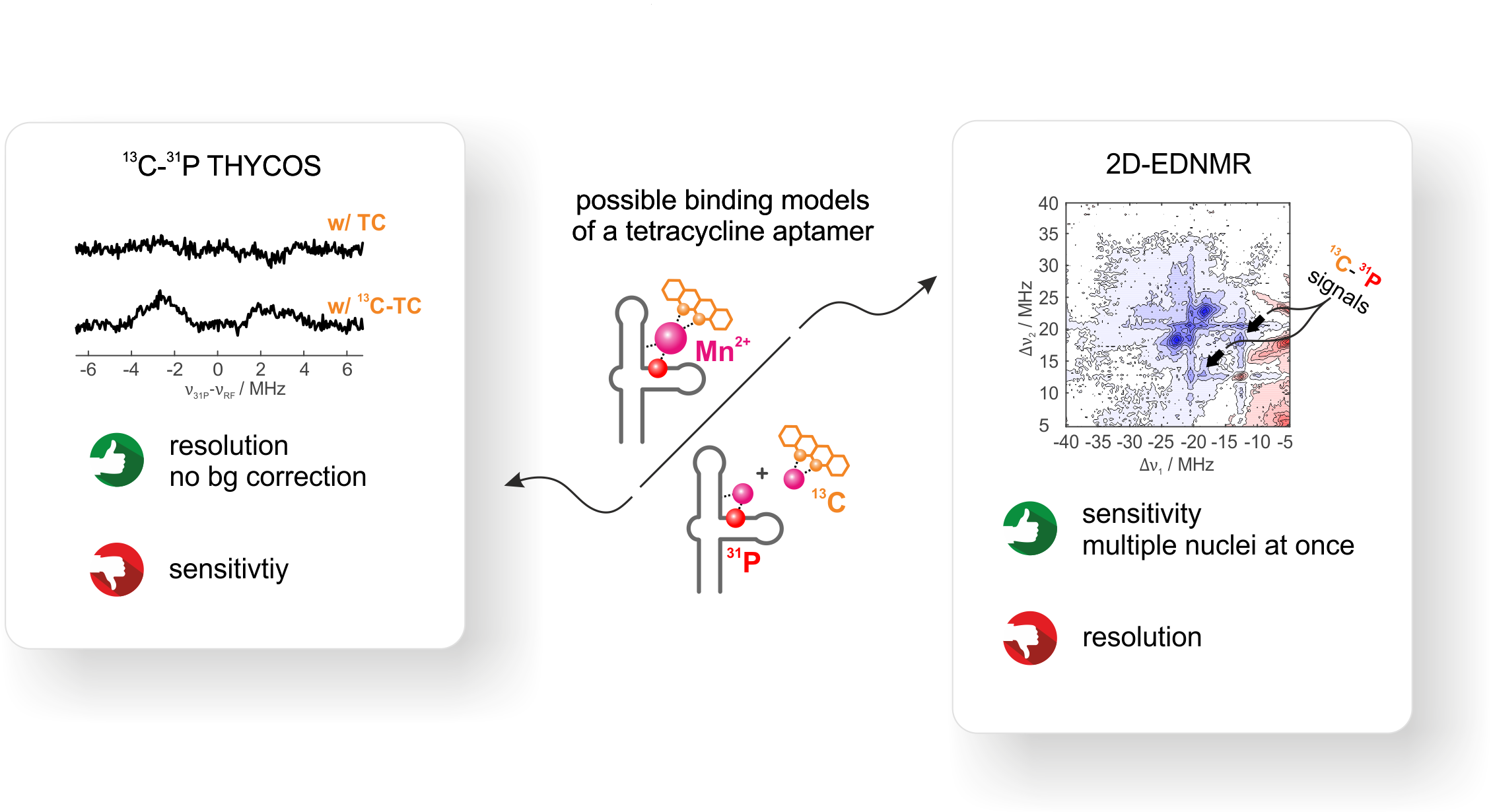
The tetracycline-binding RNA aptamer (TC-aptamer) binds its cognate ligand the antibiotic tetracycline (TC) via a Mg2+ or Mn2+ ion with high affinity
at high divalent metal ion concentrations (800 pM, ≥ 10 mM). These concentrations lie above the physiological divalent metal ion concentration of
ca. 1 mM and it is known from literature that the binding affinity decreases upon decreasing the divalent metal ion concentration.
1D-hyperfine experiments reveal two pronounced 31P couplings from the RNA besides the 13C signal of 13C-labeled TC. From these 1D-hyperfine data alone,
however, no conclusions can be drawn on the binding of TC. Either TC may bind via Mn2+ to the aptamer or TC may form a free Mn-TC complex and some Mn2+
lso binds to the aptamer.
We use THYCOS and 2D ELDOR-detected NMR at Q-band frequencies and physiological divalent metal ion concentration to correlate two different nuclear spin
species (13C and 31P) on two different molecules (RNA and TC) to the same electron spin (Mn2+).
Out of the two observed 31P-hyperfine couplings, only one shows a clear correlation to 13C.
Although THYCOS and 2D EDNMR yield identical results, 2D EDNMR is far more sensitive. THYCOS spectra needed a time factor of × 20 in comparison to 2D EDNMR
to achieve a comparable signal-to-noise.
Development of new strategies for the analysis of orientation selective PELDOR data
If rigid spin labels are incorporated into the DNA or RNA, the relative orientations between a pair of such spin labels can be extracted from
orientation selective PELDOR experiments in addition to provide their distance. This allows the determination of both the structure and the mobility
of the nucleic acid.[5] We are developing new strategies to obtain orientation information from this PELDOR data and thereby geometrical restraints
for structure determination. To improve the uniqueness of the solution for the overall structure of the nucleic acid from only a few doubly spin-labeled
mutants, we will perform orientation selective PELDOR experiments at different microwave frequencies and magnetic fields.
We have used the related spin label Ç to investigate the intrinsic conformational flexibility of dsDNA molecules [5b].
We could show in collaboration with the Hummer group of the MPI of Biophysics that such PELDOR experiments can be directly compared with MD simulations and
offer excellent benchmark data for the evaluation of force field parameters of nucleic acid molecules [6]. The quantitative comparison with
the MD simulations also confirmed that the conformational ensemble measured by EPR in the frozen state at 50 K resembles very well the dynamic conformational
freedom of the molecule at room temperature.
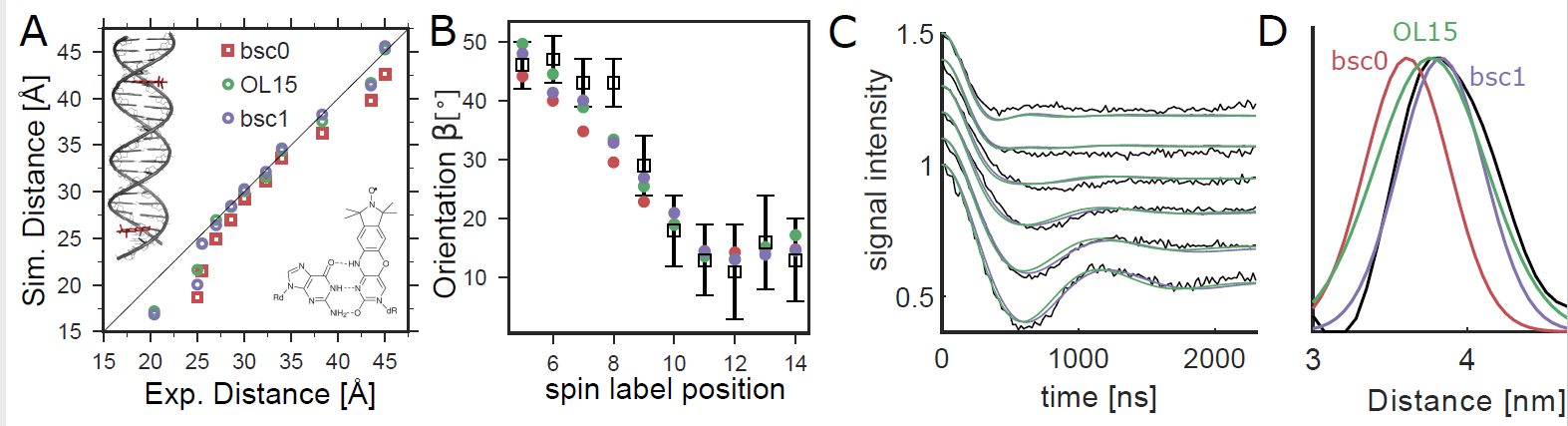
Comparison of orientation-selective PELDOR experiments (black) with MD simulations. (A) We analyzed PELDOR experiments with 11 spin-label pairs and compared distances from experiments and MD (B) Spin-label orientations from MD and PELDOR. The angle β describes the orientation between the vectors normal to the spin-label planes and the dipole−dipole vector.8 (C) Comparison of PELDOR signals calculated from MD to experiments for DNA(1,13). (D) Spin−spin distance distributions.
References:
| [1] | a) I. Krstic, B. Endeward, D. Margraf, A. Marko, T. F. Prisner, in Epr Spectroscopy: Applications in Chemistry and Biology, Vol. 321 (Eds.: M. Drescher, G. Jeschke), Springer-Verlag Berlin, Berlin," 2012, 159-198; b) I. Krstic, A. Marko, C. M. Grytz, B. Endeward, T. F. Prisner, in RNA Structure and Folding: Biophysical Techniques and Prediction Methods (Eds.: D. Klostermeier, C. Hammann), De Gruyter, 2013, 261-286. |
| [2] | Krstic, I., Frolow, O., Sezer, D., Endeward, B., Weigand, J.E., Suess, B., Engels, J.W. and Prisner, T.F., J. Am. Chem. Soc. 2010, 132, 1454-1455. doi: 10.1021/ja9077914; |
| [3] | Krstic, I., Hänsel, R., Romainczyk, O., Engels, J. W., Dötsch, V. and Prisner, T. F., Angew. Chem.-Int. Edit. 2011, 50, 5070-5074. DOI: 10.1002/anie.201100886 |
| [4] | Hetzke, T., Bowen, A.M. and Prisner, T.F., Appl. Magn. Reson. 2017, 48, 1375-1397. doi: 10.1007/s00723-017-0927-4 |
| [5] | a) Schiemann, O., Cekan, P., Margraf, D., Prisner, T. F. and Sigurdsson, S. Th., Angew. Chem.-Int. Edit. 2009, 48, 3292-3295; doi: 10.1002/ange.200805152; b) Marko, A., Denysenkov, V. P., Margraf, D., Cekan, P., Schiemann, O., Sigurdsson, S. Th. and Prisner, T. F., J. Am. Chem. Soc. 2011, 133, 13375-13379. DOI: 10.1021/ja201244u |
| [6] | Stelzl, L. S., Erlenbach, N., Heinz, M., Prisner, T. F. and Hummer, H., J. Am. Chem. Soc., 2017, 139, 11674-11677 doi: 10.1021/jacs.7b05363 |
| Last modified: February 04, 2021 | Impressum / About / Legal Notice | Datenschutzerklärung |
Prisner LOGS
Webmail
Internal
|